Time series data are widely seen in analytics. Some examples are stock indexes/prices, currency exchange rates and electrocardiogram (ECG). Traditional time series analysis focuses on smoothing, decomposition and forecasting, and there are many R functions and packages available for those purposes (see CRAN Task View: Time Series Analysis). However, classification and clustering of time series data are not readily supported by existing R functions or packages. There are many research publications on time series classification and clustering, but no R implementations are available so far as I know.
To demonstrate some possible ways for time series analysis and mining with R, I gave a talk on Time Series Analysis and Mining with R at Canberra R Users Group on 18 July
2011. It presents time series decomposition, forecasting, clustering and classification with R code examples. Some examples in the talk are presented below.
The complete slides of the talk and a comprehensive document titled R and Data Mining: Example and Case Studies are available at RDataMining website.
Time Series Decomposition
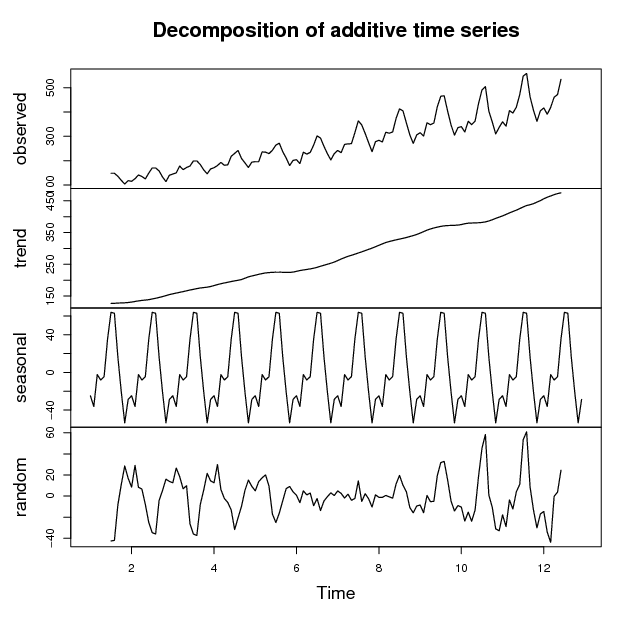
Time series decomposition is to decompose a time series into trend, seasonal, cyclical and irregular components. A time series of AirPassengers is used below as an example to demonstrate time series decomposition.
> f <- decompose(AirPassengers)
> # seasonal figures
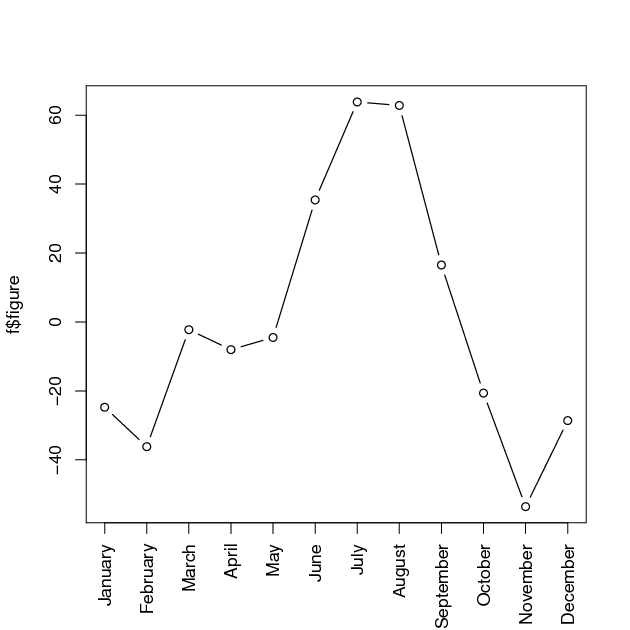
> f$figure
> plot(f$figure, type=”b”, xaxt=”n”, xlab=””)
> # get names of 12 months in English words
> monthNames <- months(ISOdate(2011,1:12,1))
> # label x-axis with month names
> # las is set to 2 for vertical label orientation
> axis(1, at=1:12, labels=monthNames, las=2)
> plot(f)
In the above figure, the first chart is the original time series, the second is trend, the third shows seasonal factors, and the last chart is the remaining component.
Some other functions for time series decomposition are stl() in package stats, decomp() in package timsac, and tsr() in package ast.
Time Series Forecasting
Time series forecasting is to forecast future events based on known past data. Below is an example for time series forecasting with an autoregressive integrated moving average (ARIMA) model.
> fit <- arima(AirPassengers, order=c(1,0,0), list(order=c(2,1,0), period=12))
> fore <- predict(fit, n.ahead=24)
> # error bounds at 95% confidence level
> U <- fore$pred + 2*fore$se
> L <- fore$pred – 2*fore$se
> ts.plot(AirPassengers, fore$pred, U, L, col=c(1,2,4,4), lty = c(1,1,2,2))
> legend(“topleft”, c(“Actual”, “Forecast”, “Error Bounds (95% Confidence)”),
+ col=c(1,2,4), lty=c(1,1,2))
Time Series Clustering
Time series clustering is to partition time series data into groups based on similarity or distance, so that time series in the same cluster are similar. For time series clustering with R, the first step is to work out an appropriate distance/similarity metric, and then, at the second step, use existing clustering techniques, such as k-means, hierarchical clustering, density-based clustering or subspace clustering, to find clustering structures.
Dynamic Time Warping (DTW) finds optimal alignment between two time series, and DTW distance is used as a distance metric in the example below. A data set of Synthetic Control Chart Time Series is used in the example, which contains 600 examples of control charts. Each control chart is a time series with 60 values. There are six classes: 1) 1-100 Normal, 2) 101-200 Cyclic, 3) 201-300 Increasing trend, 4)301-400 Decreasing trend, 5) 401-500 Upward shift, and 6) 501-600 Downward shift. The dataset is downloadable at UCI KDD Archive.
> sc <- read.table(“E:/Rtmp/synthetic_control.data”, header=F, sep=””)
# randomly sampled n cases from each class, to make it easy for plotting
> n <- 10
> s <- sample(1:100, n)
> idx <- c(s, 100+s, 200+s, 300+s, 400+s, 500+s)
> sample2 <- sc[idx,]
> observedLabels <- c(rep(1,n), rep(2,n), rep(3,n), rep(4,n), rep(5,n), rep(6,n))
# compute DTW distances
> library(dtw)
> distMatrix <- dist(sample2, method=”DTW”)
# hierarchical clustering
> hc <- hclust(distMatrix, method=”average”)
> plot(hc, labels=observedLabels, main=””)
Time Series Classification
Time series classification is to build a classification model based on labelled time series and then use the model to predict the label of unlabelled time series. The way for time series classification with R is to extract and build features from time series data first, and then apply existing classification techniques, such as SVM, k-NN, neural networks, regression and decision trees, to the feature set.
Discrete Wavelet Transform (DWT) provides a multi-resolution representation using wavelets and is used in the example below. Another popular feature extraction technique is Discrete Fourier Transform (DFT).
# extracting DWT coefficients (with Haar filter)
> library(wavelets)
> wtData <- NULL
> for (i in 1:nrow(sc)) {
+ a <- t(sc[i,])
+ wt <- dwt(a, filter=”haar”, boundary=”periodic”)
+ wtData <- rbind(wtData, unlist(c(wt@W,wt@V[[wt@level]])))
+ }
> wtData <- as.data.frame(wtData)
# set class labels into categorical values
> classId <- c(rep(“1”,100), rep(“2”,100), rep(“3”,100),
+ rep(“4”,100), rep(“5”,100), rep(“6”,100))
> wtSc <- data.frame(cbind(classId, wtData))
# build a decision tree with ctree() in package party
> library(party)
> ct <- ctree(classId ~ ., data=wtSc,
+ controls = ctree_control(minsplit=30, minbucket=10, maxdepth=5))
> pClassId <- predict(ct)
# check predicted classes against original class labels
> table(classId, pClassId)
# accuracy
> (sum(classId==pClassId)) / nrow(wtSc)
[1] 0.8716667
> plot(ct, ip_args=list(pval=FALSE), ep_args=list(digits=0))
Yanchang Zhao
RDataMining: http://www.rdatamining.com
Twitter: http://www.twitter.com/RDataMining
Group on Linkedin: http://group.rdatamining.com
Group on Google: http://group2.rdatamining.com






Pingback: Time Series Analysis and Mining with R | Share For More
Dear Yanchang,
thanks a lot for the posting. It’s very informative. However, I’m having difficulties executing the code in your second example under R 2.13 for OS X.
When executing the command:
distMatrix <- dist(sample2, method="DTW")
I get the following error message:
Error in dist(sample2, method = "DTW") : invalid distance method
Any ideas what might be going on?
Many thanks in advance,
Ruben
I think the data sample2 have some NA values, hava a look at your sample2 data!
Dear Ruben
Thank you for pointing out the problem.
I missed one statement to load library dtw before calculating distance. The statement is
library(dtw)
The package is used for dynamic time warping.
Thanks
Yanchang
RDataMining: http://www.rdatamining.com
Pingback: URL
Dear Yanchang,
Thanks for this very good work. However, just as pointed out by Reuben, I also encountered problem executing “distMatrix <- dist(sample2, method="DTW")" statement. When I changed the DTW to euclidean, it worked but the graphs are slightly different. I tried to implement the library "library(dtw)" but received this error message "Error in library(dtw) : there is no package called ‘dtw’ " . I also got the same message for library(wavelets). Any clue to these problems?
Thanks in anticipation!
Emmanuel
Hi Emmanuel
It seems that the two packages are not installed on your machine. You need to install them first before running library().
Regards
Yanchang
Dear Yanchang,
Any help on how to install these applications will be apprecaited. I guess these packages are not included in R-Studio.
Thank you.
Under rstudio, click menu “tools -> install packages”
Dear Yanchang,
Thanks for your contributions. Your suggestions have helped in solving the problem.
Dear Yanchang,
A have some time series data captured from different stations over time. My aim is to compute different in time difference between these stations for objects that matched and subsequently use the computed time to determine speed and subsequently carry out clustering and classification. I was able able to compute the time difference in seconds between two stations using the difftime function. However, in an attempt to bind the data from the respective stations, I repeatedly got error messages (such as: “Error in Ops.factor(stn12$VehicleId, stn14$VehicleId) : level sets of factors are different. In addition: Warning message:
In is.na(e1) | is.na(e2) : longer object length is not a multiple of shorter object length”)
The data contains principally of vehicleid, time stamp and station id. I have tried the following code:
stn = list.files(“H:/R/wigan”, pattern = ‘.csv’)
t12<-strptime(stn12$Date,"%d/%m/%Y %H:%M:%S") # I also tried the ts() function.
tm12<-strftime(t12,"%d/%m/%Y %H:%M:%S")
## object matching at two different stations
stn12$type = 1
stn14$type = 2
if(stn12$VehicleId==stn14$VehicleId)
d = cbind(stn12, stn14)
#unique(d$VehicleId)
#vec_ids = unique(d$VehicleId)[1:10]
vec_ids = (d$VehicleId)[1:10]
results = data.frame(start= numeric(length(vec_ids)),
end=numeric(length(vec_ids)))
i=1
for(i in seq_along(vec_ids)) {
d1 = d[d$VehicleId == vec_ids[i], ]
start = as.character(d1[1,1])
end = as.character(d1[nrow(d1),1])
results[i,] = c(start, end)
}
results
vec_ids
if(stn12$VehicleId==stn14$VehicleId)
?intersect
intersect(stn1$VehicleId,stn2$VehicleId)
merge
Any suggestions on the code for a way forward will be appreciated. Thanks for your usual cooperation.
Hope you have already fixed it.
With “
if(stn12$VehicleId==stn14$VehicleId) d = cbind(stn12, stn14)”, did you want to do a joining of two datasets in an SQL way? If yes, you might use:d <- merge(stn12, stn14, by="vehicleId")Dear Yanchang, thanks for responding. I have fixed the problem earlier. However, your suggestion is appropriate.
What is the benefit of using DWT for classification? Without pruning, I am getting 0.9516667 accuracy, and with DWT I am getting 0.8983333. I followed your code, but for my classification trees I used the following:
#classification trees
#before DWT
ctree<-ctree(ID.obs~ ., data=sc.dat)
ID.pred<-predict(ctree)
table(ID.obs, ID.pred)
(sum(ID.obs==ID.pred))/nrow(sc)
#after DWT
ctree<-ctree(ID.obs~ ., data=dwt.dat)
ID.pred<-predict(ctree)
table(ID.obs, ID.pred)
(sum(ID.obs==ID.pred))/nrow(dwt.dat)
Thanks for your comment, Johnny.
It might not produce a better result with DWT. The example is simply to demonstrate using extracted features for modelling. You might try DFT or other methods to extract potentially usefully features.
Just a followup, if we set boundary=”reflection” rather than “periodic”, we DO see a marginal benefit from DWT (0.9633 accuracy).
Several of the lines above don’t work in R, as they have the wrong character in them. (I find it necessary to turn off all the auto-format in Microsoft Word).
For example:
L <- fore$pred – 2*fore$se
That's not a minus, it's an "en-dash", so it's an error, in R. And in:
plot(f$figure, type=”b”, xaxt=”n”, xlab=”")
There are typographic quotes, instead of ", so also does not work. It's easy to fix, but you have to notice that – is not –
Thanks for the great information!
Pingback: Weekly bookmarks: januari 19th | robertsahlin.com
Hi,
thanks for this nice overview. I have also a question. The dist-function of the dtw-package returns an asymetric distance matrix, does it? Consequently, clustering is not straight forward, since on which distance (dist(a,b) vs. dist(b,a)) are the cluster-merges performed.
Or did i miss something?
Thank you!
Thanks, Julian. It is a good question. I didn’t realize that it can be asymmetric.
Regarding DTW distance in dtw package, the default setting of step.pattern is symmetric2, which produces a symmetric dist matrix.
library(dtw)mm <- matrix(runif(12), ncol=3)
## symmetirc dist matrix
dm <- dist(mm, mm, method="DTW", step=symmetric2);
dm <- dist(mm, mm, method="DTW")
However, it can be asymmetric, by setting “step=asymmetric”. In the latter case, according to the help documentation of dtwDist() in the dtw package, we can either disregard part of the pairs (as.dist), or average with the transpose.
## asymmetirc dist matrixdm <- dist(mm, mm, method="DTW", step=asymmetric);
## disregard part of the pairs
as.dist(dm)
## symmetrize by averaging
(dm + t(dm)) / 2
Thank you for pointing this out!
Dear Yanchang,
As a follow up to my previous request on how to merge data from different stations where you suggested using “d <- merge(stn12, stn14, by="vehicleId")", I would like to ask for your assistance once again.
At this stage, I am trying to work with data from many stations stored as a list which requires writing a loop that will merge all the stations together and and give the output of the individual merge data set. I have tried a couple of solutions based on my Fortran knowledge but to no avail.
Here is a brief breakdown of what I have tried:
# mdata is a list of the files to be merged
# merging more than 2 files using a loop function
mdata <- all
i<-0
i2<-0
mgr<- function(mdata){
l <- length(mdata)
for(i in mdata){
i <- i+1
i2 <- i+1
mdat[[i]] <- merge(mdata[[i]], mdata[[i2]], by = "VehicleId", sort=T,all = FALSE)[mdata[[i]],
!=mdata[[i2]]]
}
return(mdat)
}
mg <- mgr(mdata)
### another trier
i<-0
i2<-0
mdata <- all
mgr = function(mdata){
for(i in 1:length(mdata)){
i <- i+1
i2 <- i+1
mdat[[i]] <- merge(mdata[[i]], mdata[[i2]], by = "VehicleId", sort=TRUE,
all = FALSE)
}
return(mdat[[i]])
}
mg <- lapply(mdata,mgr)
However, if I run the following for any combination of two files, it will give me the output as desired.
mdat <- merge(mdata[[1]], mdata[[2]], by = "VehicleId", sort=TRUE,all = FALSE)
Any help will be highly appreciated. Thank you for your usual cooperation
Sorry, Emmanuel, for my late reply. I don’t check this blog very often. For future questions, please post them to my RDataMining group on LinkedIn at http://group.rdatamining.com/, which I check everyday.
Please let me know if you still haven’t solved above problem.
Dear Yanchang, thanks for finding time to reply and to let me know the active blog. I have solved the problem. Many thanks!
I
have not solved the problem yet
Pingback: Dynamic Time Warping using rpy and Python |
i want to work on data mining in time series data using R, please what are the steps to take to accomplish the project
Hi Tosin, your question is too general. What is the problem you want to solve in the project? Forecasting or something else?
I’d like to suggest you to post your questions to my RDataMining group on LinkedIn at http://group.rdatamining.com/. Thanks.
Hello, can you explain me how to get the ast package? could it be removed?
See the package at http://sirio.stat.unipd.it/index.php?id=libast. The page is not in English, but you can read it with Google Translate.
Hi,
Do you perform any kind of scaling or anyting on original data, before you do DWT for classification. I have some time series (financial) of stocks, each has a different price. I changes all the time series to be Yield, so they are all kind of same scale. Still not getting good classification. So trying to get any suggestion from you on pre process of time series for classification
Pingback: 선물거래와 Dynamic Time Warping
Hello Yanchang Zhao,
I see that you have used ARIMA model in Time Series Forecasting section, Can you also please share some thoughts on how do we perform “seasonal linear regression” on the time series data.
ie. instead of directly using the ARIMA model is there any other function that can be used to evaluate the series for the seasonality and if it exhibits the seasonality then a seasonal linear model should forecast the same.
Thanks.
Harshad M.